The Tangled Tree, David Quammen (2019)
I have just read David Quammen’s The Tangled Tree: A Radical New History of Life (2019). It is a beautifully written book on molecular phylogenetics. Quammen has written over a dozen books on the life sciences, and he is a great storyteller and science journalist.
I recommend this book, with one serious reservation. It describes a purely evolutionary view of molecular phylogenetics. Quammen unfortunately entirely ignores convergent evolution, and thus never allows the reader to consider its implications for universal development. He also does not discuss evo-devo biology. If he had, he might have recognized just how constraining accretive processes of biological development must be on all macrobiological evolutionary change.
Consider the fact that all complex animals, including humans, share almost all the same basic developmental regulatory machinery found in much simpler organisms than us. Like a tree that grows outward from a central trunk, we can’t update our developmental code as we grow more complex. We can only add to that code, progressively limiting our morphological and functional options in evolution. Constraining factors like accretive regulatory development and convergent evolution are physical realities we must recognize if we are to understand long-range macrobiological change on Earthlike planets.

Convergent evolution in antifreeze proteins in Arctic and Antarctic fish.
Scientists have been researching the molecular phylogenetics of convergent evolution since the 1990s, when evo-devo biology first became a formal subdiscipline. For example, we’ve known since 1997 that antifreeze proteins evolved via two clearly independent genetic means in Northern and Southern polar fish, to prevent ice crystal formation.
As our science and simulation advance, I think we will discover a vast number of developmental portals, uniquely adaptive and accelerative attractors on the road to competitive complexification that must be discovered via evolutionary search in our universe. Such complexification attractors have long been proposed by developmentally-oriented thinkers. Organic chemistry, Earthlike planets, nucleic acid-, protein-, and fat-based cells, oxidative phosphorylation, multicellularity, nutrient- and waste-carrying circulatory systems, and the emergence of antifreeze in animals living in near-zero temperature habitats are just a few of many proposed examples of such adaptive attractors. I’d argue they are examples of what EDU scholar Claudio Flores Martinez calls “cosmic convergent evolution” [SETI in the light of cosmic convergent evolution, Acta Astronautica, 104(1):341–349, 2014].
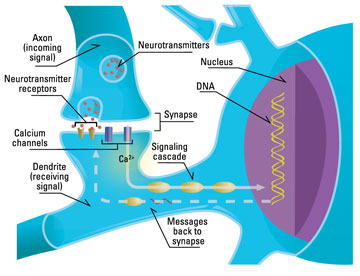
Fortunately, we can increasingly investigate some of the more recent proposed attractors via molecular phylogenetics, inferring the recent genetic history of life on Earth. Some of these more recent attractors include nervous systems, which according to Flores Martinez appear to have independently emerged at least three times (in bilaterians like us, in comb jellies, and in jellyfish) using three different neurotransmitter schemes. If nervous systems are a true portal, there won’t be anything else that can be built on top of our kind of multicellularity that would give collectives a comparable competitive advantage. In bilaterians, emergences like endoskeletons, muscles, prehensile limbs, opposable thumbs, emotions, ethics, language, consciousness, and extrabiological tool use have all been proposed as additional portals that are uniquely able to support accelerating complexification in collectives in their local environments. Such universal developmental checkpoints, if they exist, must be reliably statistically accessible, dominant, and persistent when discovered via evolutionary search. Today, increasing numbers of proposed universal adaptive convergences are becoming accessible to molecular investigation.
With respect to antifreeze in polar environments we learned in the 1990s that the antifreeze gene used by a Southern fish, Antarctic cod, arose from a mutation of gene that originally coded for a digestive enzyme. But the origin of the antifreeze protein in the Northern polar fish, Arctic cod, remained unclear. This 2019 article by Ed Yong at The Atlantic describes how, after twenty more years of diligent work, Chi-Hing Christina Cheng and her group deduced the complex way that Arctic fish built their antifreeze protein. It arose from a stretch of noncoding DNA, which was duplicated, mutated, relocated next to a promoter, and then a base was deleted to make it functional. In the twentieth century, some geneticists used to think noncoding DNA was “junk”. Work like Cheng’s tells us that noncoding DNA offers life a deep pool of potential genetic and protein diversity. We’ve also found antifreeze (and many other wintering adaptations) in other cold-dwelling species, like Cucujus clavipes, the red bark beetle. I’m sure we’ll learn many more stories of convergence there as well.
If Quammen had recognized that convergent molecular phylogenetics offers an exciting new way to understand long-known morphological and functional convergence in phylogenetically unique species, just as molecular methods give us exciting new ways to understand phylogenetics, he would have done a great service to general readers and scholars alike. Morphological and functional convergences, along with some hints at genetic and molecular evo-devo pathways toward them, have long been described by scientists like Simon Conway Morris (Life’s Solution, 2004; The Deep Structure of Biology, 2008), Johnathan Losos (Improbable Destinies: How Predictable is Evolution?, 2018, and George McGhee (Convergent Evolution: Limited Forms Most Beautiful, 2011; Convergent Evolution on Earth, 2019).
Work like this tells us that our morphological and functional tree of life (a separate concept from our phylogenetic tree) is both continually diverging, due to contingent evolutionary innovation, and continually converging, due to the existence of universal environmental optima that will inevitably discovered, on all planets with environments like ours, via evolutionary search. In important ways then, this latter tree of life is significantly less tangled than it first seems. Life, a macrobiological system with fixed and finite complexity, is going somewhere, developmentally speaking. Both evolutionary contingency and developmental inevitability are central to the story of life on Earth, and other Earthlike planets in our universe.
We started our Evo Devo Universe (EDU) research and discussion community in 2008 precisely because the story of universal development is so widely ignored and downplayed. Most scientific work today perpetuates the one-sided, evolution-only view of change and selection that is the dominant scientific narrative today. There seems to be a strong emotional commitment among some scientists to the idea of an almost entirely contingent universe. Perhaps this commitment arises because of the unsettling implications of a universe that is developing as well as evolving. If our universe is developmental, science may become not merely descriptive, but prescriptive. It may learn to tell how we may better act, to be in service to universal processes and goals.
My new paper, Evolutionary Development: A Universal Perspective (2019) is my own latest small effort to offer an opposing, evolutionary developmental perspective. For a lay article on why we appear to live in an evo-devo universe, you may enjoy my post Humanity Rising: Why Evolutionary Development Will Inherit the Future (2012).
One of the books high points is its excellent discussion of the great Carl Woese. Woese and his student, George Fox, revolutionized microbiology by realizing we could trace bacterial phylogenetics through internal “molecular fossils.” They deduced the phylogenetic taxonomy of 16S ribosomal RNA, the universal machinery of protein manufacturing. This work allowed them to classify Archea, single-celled organisms that have a more complex internal structure than bacteria. Archaea range widely on Earth, and engage in a great variety of energy metabolisms (sugars, ammonia, metal ions, hydrogen gas), unlike their simpler bacterial cousins.

Woese and Fox’s Tree of Life, 1977
Woese’s work gave us our modern phylogenetic tree of life in 1977 (picture right). This tree showed that Archea are closer in phylogenetic history to us than bacteria. It is a good bet that both eukaryotes and prokaryotes branched off from an Archea that lived in undersea geothermal vents, making energy from hydrogen gas, warm water, and underwater nutrients richly available in those vents. Chemosynthesis, in other words, likely arrived on Earth long before photosynthesis.
What’s more, life on Earth appears to have emerged almost as soon as our planet became cool enough to support liquid water. Metal-rich Earthlike planets, with plate tectonics, plentiful water, and volcanic vents, appear to be ideal catalysts for life, and our geochemical cycles are ideal buffers and cradles for stabilizing life once it emerges. The complex set of homoeostatic protections for life on Earth, aka the Gaia hypothesis, when stated without the woo of “planetary intelligence”, appear far more developmental, from a universal perspective, than the hypothesis’s many detractors like to admit.
Woese’s work also lends credence to Alexander Rich and Walter Gilbert’s RNA world hypothesis, the idea that self-replicating RNA emerged first, before DNA and proteins. RNA is one of those rare complex chemicals that can store memory of its past evolutionary variation and self-catalyze its own replication. In other words, it is autopoetic (capable of self-maintenance and self-improvement).
Another high point is the book’s discussion of horizontal gene transfer. Amazingly, it appears that about 8% of human DNA arrived sideways in our genome, not via sex or mutation but via viral infection. As Harald Brüssow reminds us in “The not so universal tree of life,” we have not yet incorporated viruses into our current trees of life. That is a major oversight. Retroviral insertion sequences are found everywhere in eukaryotic DNA. Viruses and cells are constantly exchanging genetic material, in all species. [Brüssow H. (2009). The not so universal tree of life or the place of viruses in the living world. Phil trans. Royal Soc. of London. doi:10.1098/rstb.2009.0036]

Tree of life showing vertical and (a few) horizontal gene transfers. Source: Wikipedia
Our Real Tree of Life, once we draw it to include viruses, will look even more like a network than in the figure at right. The tree drawn at right is a good step beyond Woese’s 1977 tree, but it is still much too conservative. It includes no lines between eukaryotes, for example. It ignores retroviruses and other mechanisms. See the Wikipedia article on HGT for the great variety of DNA transfer mechanisms we’ve discovered so far.
DNA is arguably still the dominant autopoetic system on our planet today. DNA’s astonishing ability to copy, vary, and improve itself, to jump around inside the cell as transposons, to jump between cells and organisms via viral and retroviral insertion, and to use vertical methods like germline mutation and sexual recombination, has made all living species on Earth much more of a single interdependent network than most of us realize.
This is an important idea to understand, because is the genetic network, not any collection of species, that has always been the true survivor and improver in life’s story. Many past environmental catastrophes, like the Permian extinction, and the K-T meteorite impact, have wiped out the vast majority of species, but I would personally bet almost all of the diversity of the genetic network survived each of those events. This is obviously true in developmental genes, which are highly conserved. If any complex species survives a catastrophe, the developmental core of all complex species survives. But I suspect it is true for most evolutionary (nonconserved) genes as well. We shall see if the evidence from modern catastrophes bears this assertion out. Genes are typically reassorted into hardier species after each catastrophe, and those species, having no competition and ample resources, make great leaps in innovation immediately after each major catastrophe. I call that the catalytic catastrophe hypothesis, and I look forward to seeing it proven in coming years.
Interdependent networks, in other words, always win out in complex selective environments, over time. Such networks are stabler, safer, more ethical, and more capable than isolated individuals. There are deep lessons in complexity science and network science to be discovered here, lessons that tell us why our leading forms of artificial intelligence later this century will be driven to not only be deeply biologically-inspired, but also ethical, empathic, and self-regulating collectives, just like us. Complex selection and developmental optima will ensure this is so, statistically speaking, in my view.
Again, if Quammen had covered convergent molecular phyogenetics, and a bit of evo-devo and developmental genetics, he would he would have given us a better set of trees and networks to ponder. If he’d wrestled with the convergent features of biological development at the organismic scale, he might have begun to recognize it at the ecosystem scale, and help us to begin to see and ponder it too.
Life is a complex, interdependent network, but it is also going somewhere. It is developing, not just evolving. I speculate on the intrinsic goals of evo-devo systems in my 2019 paper above. It may be too early to for us to say with certainty what goals life has, as a complex evo-devo network, but it is not to early to recognize that such goals must exist, both from evolutionary and developmental perspectives.
When considered as a single interdependent network, life’s story on Earth so far has been a curiously smooth and continually accelerating trajectory of increasing complexity, stability, ability, and intelligence. Something very curious is going on in all the Earthlike, high-complexity environments in the universe. We need to start recognizing and studying it much more closely if we wish to understand accelerating change, complexity and adaptation from a universal perspective, not just our own.






 Until
Until